NucleoMag NGS Clean-up and Size Select
NucleoMag NGS Clean-up and Size Select
Applications
- Clean up / size selection of NGS library preparation reactions
Features
- Easily adjustable for different applications or workflows
- Tunable size selection from 150 bp to 800 bp – highest flexibility for customer specific applications
- Magnetic bead technology allows scalability in manual and automated workflows
Specifications
- Technology: Magnetic bead technology
- Processing: Manual or automated
- Sample material: Reaction mixtures from NGS library kits
- Amount of sample material: 7.5 pg–5 μg
- Fragment size: 150–800 bp (tunable)
- Input volume: 50–150 μL
- Typical recovery: > 80 %
- Elution volume: 10–100 μL
- Processing time: 45–60 min/96 preps
High recoveries in NGS library preparation
Most sequencing platforms for library preparations require enzymatic reaction clean-up and fragment size selection. NucleoMag® NGS Clean-up and Size Select enables clean-up reactions, as well as a single or double side size selection, to recover the fragment lengths which are needed in your specific application. The magnetic beads in the kit are present in binding buffer and just have to be diluted to tune the required fragment size. The dilution ratio is similar to competitive beads in the market. This allows the NucleoMag® NGS Clean-up and Size Select kit to be used with your current protocols without the need to make any changes.
Size selection of fragment mix
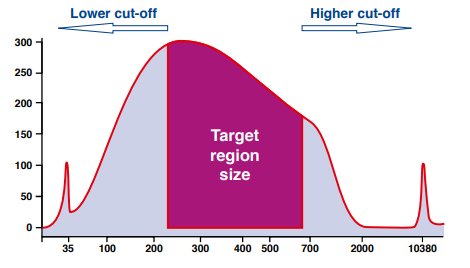
For single side size selection (left or right), the sample is mixed with the beads in predetermined ratios for the desired exclusion of smaller or larger sized fragments. For the double sized size selection, two binding steps are performed to exclude larger fragments above the cut-off and smaller fragments below the lower cut-off.
Fragment size analysis of prepared NGS libraries
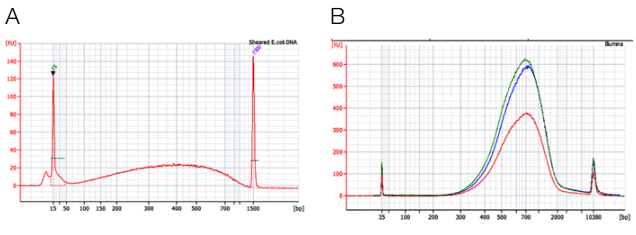
NGS libraries were prepared using Truseq™ DNA PCR Free kit and Illumina (red), AMPure® XP (blue) and NucleoMag® NGS Clean-up and Size Select (green) for clean-up and size selection steps. (A) Input DNA, 1 μg sheared E. coli DNA. (B) Size distribution of DNA Fragments after library preparation as input for sequencing, the expected fragment size is 650 bp (insert + adapters).
Sequencing analysis
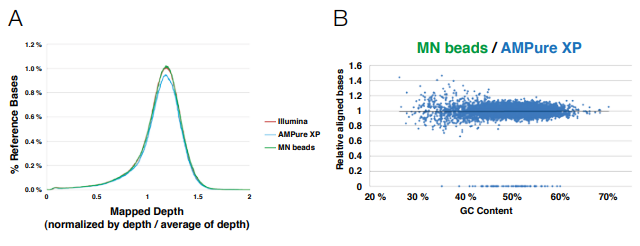
In order to compare the sequencing performance after library preparation generation with different clean-up and size selection products, the distribution of genome sequence coverage (depth of genome base coverage) and effect on GC content on sequence coverage was tested. (A) No difference in mapped depth of genome base coverage. The depth is constant indicates low bias. (B) Horizontal plot pattern of MN beads against Agencourt beads. After the summary of aligned read bases within a 0.5 kb genome region, the two samples have been normalized / matched resulting in a horizontally pattern showing the similarity of the two samples
Application data
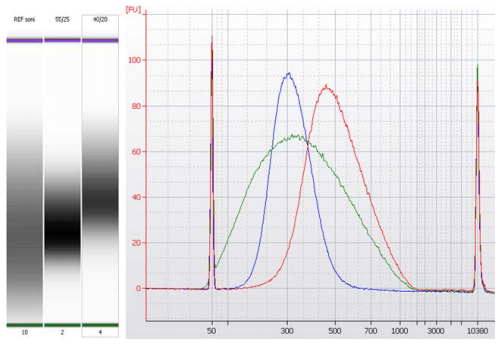
Many applications for DNA analysis (especially in the field of NGS) require a finely tuned size of DNA fragments. This is most precisely achieved by double size selection. In short, the NGS beads are mixed with the sample of interest in a ratio that allows for selective binding of fragments larger than the size of the fragment size range of interest (right side selection). Afterwards, this first batch of beads with the bound, unwanted DNA is discarded and fresh beads are added in a ratio that allows for binding of the fragment of choice (left size selection). The smaller DNA fragments are discarded with the supernatant and the DNA of interest is washed and eluted from the beads. In this experiment, total mouse tissue DNA was subjected to shearing, creating a broad range of fragment sizes (green curve). This mix was afterwards subjected to two different double-size selection procedures, a right 0.4 ratio / left 0.6 ratio pair selecting for fragments sizes of 460 bp (red peak) and a right 0.55 / left 0.8 pair selecting for 240 bp (blue peak), respectively. Many more ratio pairs are possible, allowing for size selection of other fragment sizes.
green: DNA fragment size distribution from mouse tissue after fragmentation without size selection
red: DNA fragment size distribution after double sided size selection with dilution ratios of 0.4 (right) and 0.6 (left); mean fragment size: 460 bp
blue: DNA fragment size distribution after double sided size selection with dilution ratios of 0.55 (right) and 0.8 (left); mean fragment size: 340 bp