SMART-Seq Single Cell Kit—full-length transcriptome analysis with ultimate sensitivity for single cells
The SMART-Seq Single Cell Kit (SSsc) is powered by robust chemistry that provides unparalleled sensitivity and reproducibility for single-cell and nuclei applications. This kit uses oligo(dT) priming to generate high-quality, full-length cDNA directly from single cells known to have low RNA content (e.g., PBMCs, T-cells, B-cells, etc.).
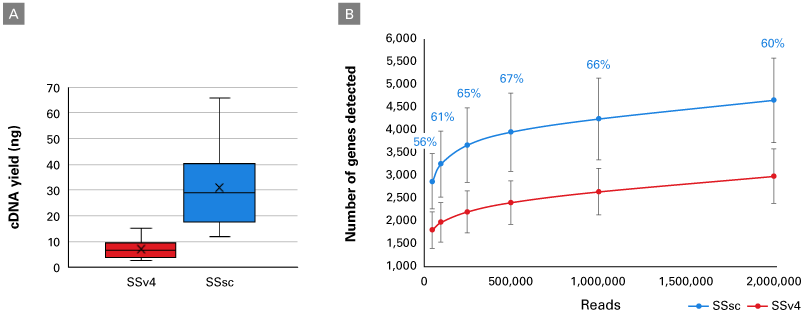
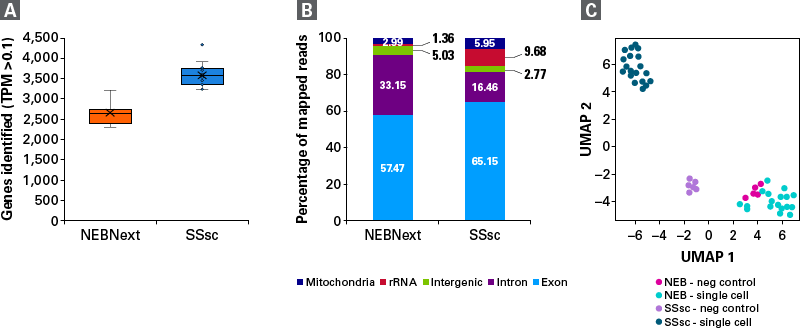
The SMART (Switching Mechanism at 5′ End of RNA Template) technology powering the SMART-Seq Single Cell kit provides full-length transcript information, enabling analysis of transcript isoforms, gene fusions, point mutations, etc. The cDNA kit provides high reproducibility, even gene-body coverage, and an accurate representation of GC-rich transcripts. We have further modified this powerful core technology to create a new chemistry with higher sensitivity designed specifically for single-cell applications. These enhancements result in more useful reads and a higher gene detection, allowing for a better understanding of precious samples.
The SSsc PLUS kit includes our patented library preparation kit that incorporates enzymatic fragmentation and stem-loop adapters to construct high-quality, Illumina-compatible libraries from the cDNA synthesized with the core kit. This two-step workflow takes place in a single tube and can be completed in about two hours with no intermediate purification steps necessary—minimizing sample mix-up, sample loss, and cross-contamination. The high library yield generated from this kit provides the flexibility to sequence on high-throughput sequencers while providing high reproducibility.
If you are interested in these benefits but are working with intact, high-quality cells (<1,000) or ultra-low total RNA inputs, we recommend our SMART-Seq v4 Kit.
If you prefer a random priming approach that will allow you to work with degraded samples and includes library preparation and indexing reagents, we recommend our SMART-Seq Stranded Kit.
Overview
- Easy, plate-based workflow—direct input of single cells isolated by FACS or other methods
- Unparalleled sensitivity and reproducibility—get the best performance for single-cell RNA-seq (especially for cells with very low RNA content and for nuclei) with the highest gene detection
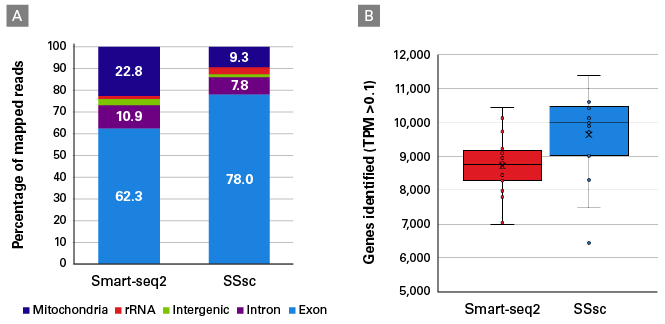
- Robust, full-length chemistry—outperform all existing single-cell full-length methods, including Smart-seq2, with this validated protocol
- Increased sequencing power—include unique dual indexes to allow for the pooling of multiple samples and confident sequencing on the NovaSeq™ system
- Highly scalable workflow—automate and use for high-throughput applications
Applications
- cDNA synthesis from single cells for full-length transcriptome sequencing
- cDNA outputs from the SSsc kit can be used with the SMART-Seq Library Prep kit (not sold separately; sold as part of the SSsc PLUS kit) for Illumina sequencing