Full transcriptome analysis of single cells with the SMART-Seq Stranded Kit
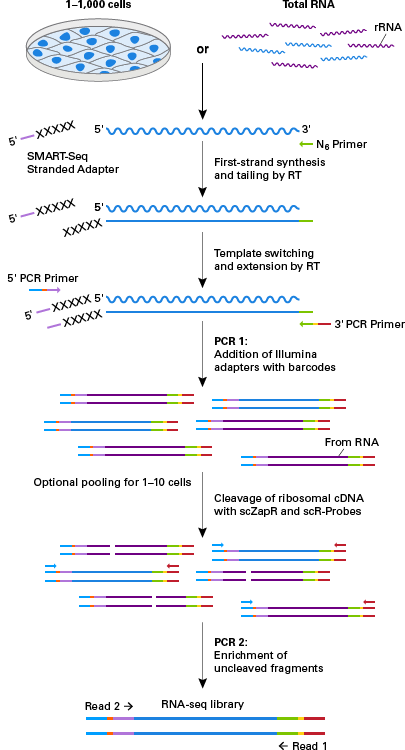
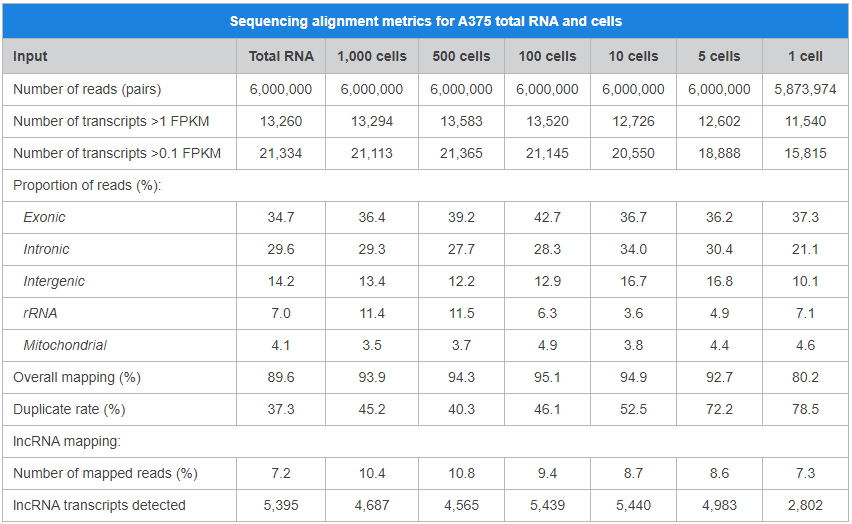
The SMART-Seq Stranded Kit is used to generate strand-specific RNA-seq libraries for Illumina® sequencing from 1–1,000 sorted cells or 10 pg–10 ng of purified total RNA. This kit incorporates Takara Bio’s SMART (Switching Mechanism at the 5′ end of RNA Template) technology and includes refinements to the SMARTer method for stranded RNA-seq that simplify the library preparation workflow and improve sequencing performance. In addition, contrary to the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing and SMART-Seq HT Kit, reverse transcription is initiated using random priming instead of oligo(dT) priming, thus capturing the full transcriptome instead of only the polyadenylated fraction. The SMART-Seq Stranded Kit was specifically designed to deliver highly sensitive and reproducible data from single cells while keeping the workflow short and user friendly. The kit does not require additional rRNA removal methods or kits and produces sequencing libraries that retain strand-of-origin information. The integrated removal of cDNAs derived from rRNA-typically present in high abundance following cDNA synthesis from total RNA inputs-makes the workflow extremely sensitive, yielding data that is highly reproducible with low mapping to rRNA.
Overview
- Simple workflow to generate stranded Illumina sequencing-ready libraries in 7 hours
- Appropriate for analysis of 1–1,000 intact mammalian cells or 10 pg–10 ng of low- or high-quality mammalian total RNA
- Reproducible, accurate detection of full-length coding and noncoding transcripts from single cells or total RNA
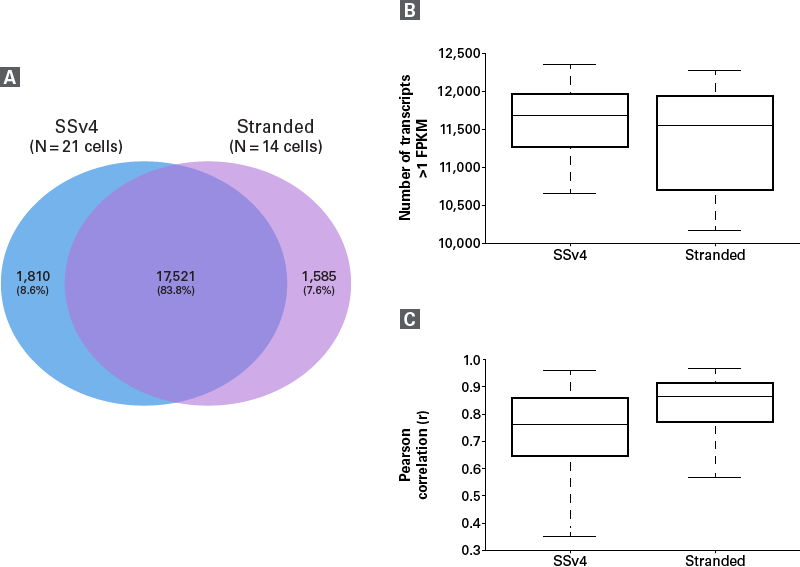
- Comparable sensitivity to our gold-standard SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing.
Applications
- Generation of Illumina sequencing-ready libraries in 7 hours from small quantities of mammalian cells (1–1,000 cells) or total RNA (10 pg–10 ng)
- Whole transcriptome sequencing with strand-of-origin information