SMARTer smRNA-Seq Kit for Illumina—sequence small RNAs with high sensitivity and minimal bias
Discovery and analysis of small non-coding RNAs (smRNAs) has become an important part of understanding gene expression regulation. Some of the well-known small RNA species are miRNAs, siRNAs, piRNAs, and snoRNAs, many of which range in size from 20 to 30 nt. The SMARTer smRNA-Seq Kit for Illumina has been specifically designed to generate high-quality Illumina-ready sequencing libraries from low-input amounts of small RNAs such as these.
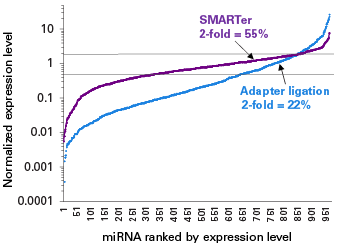
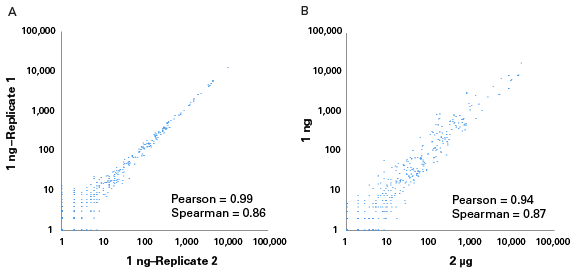
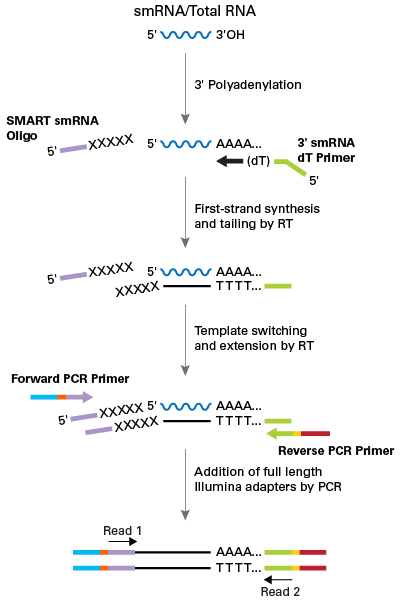
This kit was developed to work directly from 1 ng–2 µg inputs of total RNA or enriched small RNA samples, and incorporates features from the industry-leading SMART-Seq v4 kit, including SMART technology (Switching Mechanism at 5’ End of RNA Template) and locked nucleic acids (LNAs). Libraries are generated in a ligation-free manner, ensuring that diverse small RNA species are represented with minimal bias.
Overview
- Analysis of diverse small RNA species—Sequencing library prep of small RNAs ranging in size from 15–150 nt
- Flexible sample inputs—Start from 1 ng–2 µg of total RNA or enriched small RNA
- Illumina-ready sequencing libraries—Ligation-free incorporation of Illumina adapters and indexes for sample multiplexing
- Quick, single-tube workflow—Generate sequencing libraries in ~3 hours (not including validation and post-PCR size-selection steps)
Applications
- Illumina-ready sequencing library preparation, specifically for small RNAs ranging in size from 15–150 nt