SMARTer Mouse BCR IgG H/K/L Profiling Kit
SMARTer Mouse BCR IgG H/K/L Profiling Kit
SMARTer Mouse BCR IgG H/K/L Profiling Kit—capturing complete V(D)J variable regions of BCR transcripts
The SMARTer Mouse BCR IgG H/K/L Profiling Kit pairs 5' RACE with NGS technology to provide a sensitive, accurate, and optimized approach to BCR profiling. The 5'-RACE method reduces variability and allows for priming from the constant region of BCR heavy or light chains. This kit combines these benefits with gene-specific amplification to capture complete V(D)J variable regions of BCR transcripts and provide a highly sensitive and reproducible method for profiling B-cell repertoires.
The kit is designed to work with a range of RNA input amounts depending on the sample type, and has been shown to generate high-quality, Illumina-ready libraries from as little as 10 ng to 3 µg of total RNA from spleen, lymph node, peripheral blood mononuclear cells (PBMCs), and hybridomas.
Downstream sequencing of H/K/L chains allows for accurate identification of top clonotypes and reliable assignment of isotype in a majority of cases.
Overview
- Input: 10 ng–3 µg of total RNA from spleen, lymph node, PBMCs, and hybridomas
- Sensitive and specific clonotype detection with optimized cDNA library generation
- Accurate amplification of mouse IgG subclasses and identification via sequencing in a majority of cases
- Optimized PCR pooling strategy for highly sensitive detection of different chains from the same sample
- Flexible pooling strategies accommodate different experimental requirements regarding alignment and identification of primary chain sequences
Applications
- Mouse BCR repertoire analysis (heavy and light [kappa and lambda] chains)
Development of B-cell receptors

BCR development. The progenitor cell undergoes recombination of V, D, and J segments in the germline, which generates two identical heavy chains. Recombination of V and J segments generates two identical light chains. Random nucleotide additions or deletions at the junctions of the V, D, and J segments provide additional diversity. Furthermore, B cells activated by immune responses undergo somatic hypermutation (SHM), in which additional point mutations are introduced.
SMARTer Mouse BCR IgG H/K/L Profiling Kit workflow
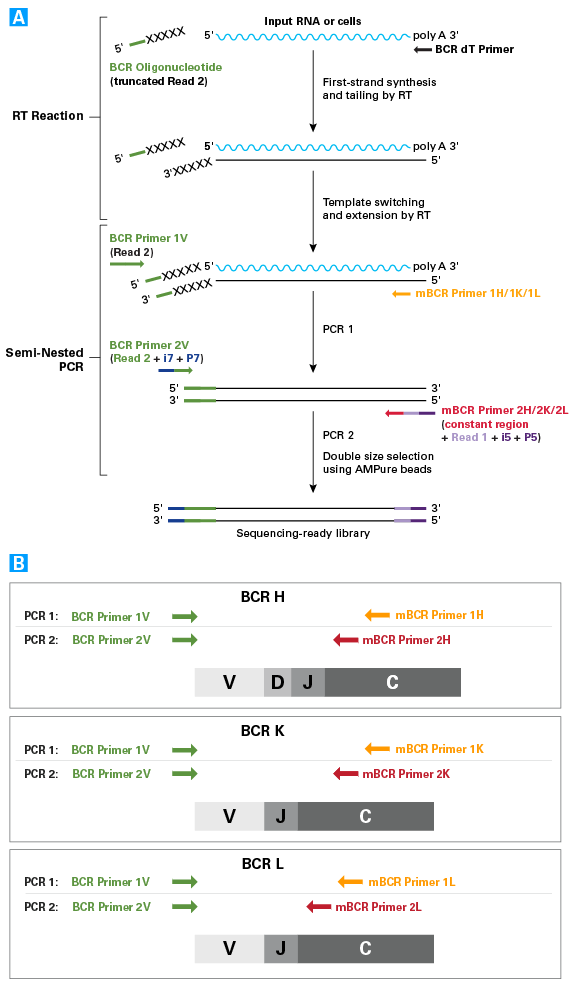
SMARTer Mouse BCR IgG H/K/L Profiling Kit workflow. Panel A. First-strand cDNA synthesis is dT-primed (BCR dT Primer) and performed by the MMLV-derived SMARTScribe Reverse Transcriptase (RT), which adds nontemplated nucleotides upon reaching the 5' end of each mRNA template. The BCR Oligonucleotide anneals to these nontemplated nucleotides and serves as a template for the incorporation of an additional sequence of nucleotides into the first-strand cDNA by the RT (this is the template-switching step). The BCR Oligonucleotide contains sequence from the Illumina Read Primer 2, serving as a primer-annealing site for subsequent rounds of PCR, and ensuring that only sequences from full-length cDNAs undergo amplification. Panel B. The first PCR uses the first-strand cDNA as a template and includes a forward primer with complementarity to the Illumina Read Primer 2 sequence (BCR Primer 1V), and a reverse primer that is complementary to the constant (i.e., nonvariable) region of BCR heavy or light chains (mBCR Primers 1H, 1K, or 1L). The chains are amplified in separate reactions. By priming from the Read Primer 2 sequence and the constant region, the first PCR specifically amplifies the entire variable region and a considerable portion of the constant region of BCR heavy or light chain cDNA. The second PCR takes the product from the first PCR as a template and uses semi-nested primers (mBCR Primers 2H, 2K, or 2L) to amplify the entire variable region and a portion of the constant region of BCR heavy or light chain cDNA. As in PCR 1, the BCR subunit chains are amplified in separate reactions.
Specific amplification of IgG subclasses and kappa and lambda light chains

Accurate amplification of all five subclasses of IgG. Libraries containing BCR heavy and light chain (kappa) sequences were generated using the SMARTer Mouse BCR IgG H/K/L Profiling kit, starting with 10 ng of RNA isolated from an in-house hybridoma (10E8) and two ATCC hybridoma samples (HB-8117 and TIB-127) with different IgG subtypes. Panel A. Bioanalyzer traces show gene-specific amplification of heavy or light chains of each hybridoma. In each sample, the expected peak between 700–900 bp is observed. The strong discrete peaks in the electropherograms show that the kappa chain is being expressed in these hybridomas. In contrast, the lambda chain sequencing library is comparatively broad and of a smaller size range than expected. Peaks labeled "LM" and "UM" correspond to DNA reference markers included in each analysis. Panel B. Mapping metrics determined using MiXCR software (version 1.8), aligned against all IG sequences. Output from the MiXCR software for the top two clones shows the V, D, and J alleles and the isotype and subclass as identified by the software.
Flexibility in PCR pooling for different experimental requirements

PCR pooling affects alignment and on-target percentages. Libraries containing BCR heavy and light chain sequences were generated using the SMART Mouse BCR kit as described in the user manual. If only heavy and kappa light chain PCR products are pooled and sequenced ("HK" in the table), a high percentage of the sequences align to IG reference sequences (using MiXCR software version 1.8) for each sample. If the lambda light chain PCR products are also pooled ("HKL" in the table), alignment percentages drop. However, the majority heavy chain and kappa chain sequences (top two clones) are identical and in the same proportion for each sequencing sample. CDR3 amino acid sequence, which defines the affinity of the antibody, is given for each clonotype shown.


