SMARTer Human scTCR a/b Profiling Kit
SMARTer Human scTCR a/b Profiling Kit
SMARTer Human scTCR a/b Profiling Kit—elucidate alpha-beta chain pairings of T-cell receptors in single cells
Although sequencing of bulk T cell samples has helped to further understanding of T-cell receptor (TCR) repertoire diversity, bulk sequencing cannot determine the pairing of specific alpha-beta receptor chains within the population of T cells. Since the unique alpha-beta pairing of a TCR mediates antigen specificity, obtaining pairing information will be crucial for providing insights on antigen recognition. While sequencing of single T cells can determine the pairing of a given cell, it can be prohibitively expensive and involves complicated analyses since most methods require sequencing of a large number of cells.
With the SMARTer Human scTCR a/b Profiling Kit, we solve this problem by performing SMART cDNA synthesis with a set of unique indexed oligos that allow the condensing of 96 samples, which have been sorted manually or via FACS, into twelve pools for sequencing. Additionally, the approach enables the twelve pools to be further multiplexed such that all 96 samples are run in a single flow-cell lane. Much like the SMARTer Human TCR a/b Profiling Kit for bulk samples, this kit combines SMART cDNA synthesis and RACE-based PCR, followed by TCR gene-specific PCR to fully capture and amplify TCR-alpha and TCR-beta variable regions to generate Illumina-ready libraries that provide a highly sensitive approach to sequencing TCRs.
Overview
- Flexible workflow: Illumina-ready libraries from FACS or manually sorted single cells
- Ease of use: optimized indexing allows for pooling 96 cells into twelve libraries which can be further multiplexed for running in a single flow-cell lane
- Sensitivity: RACE-based approach allows for the detection of low-abundance TCR variants
- Specificity: full-length reads, with the majority of reads on target and accurate pairing information
Applications
- Human TCR repertoire analysis in single cells (TCR-alpha and TCR-beta subunits)
- TCR alpha-beta pairing determination in single cells
Why single-cell TCR analysis is needed
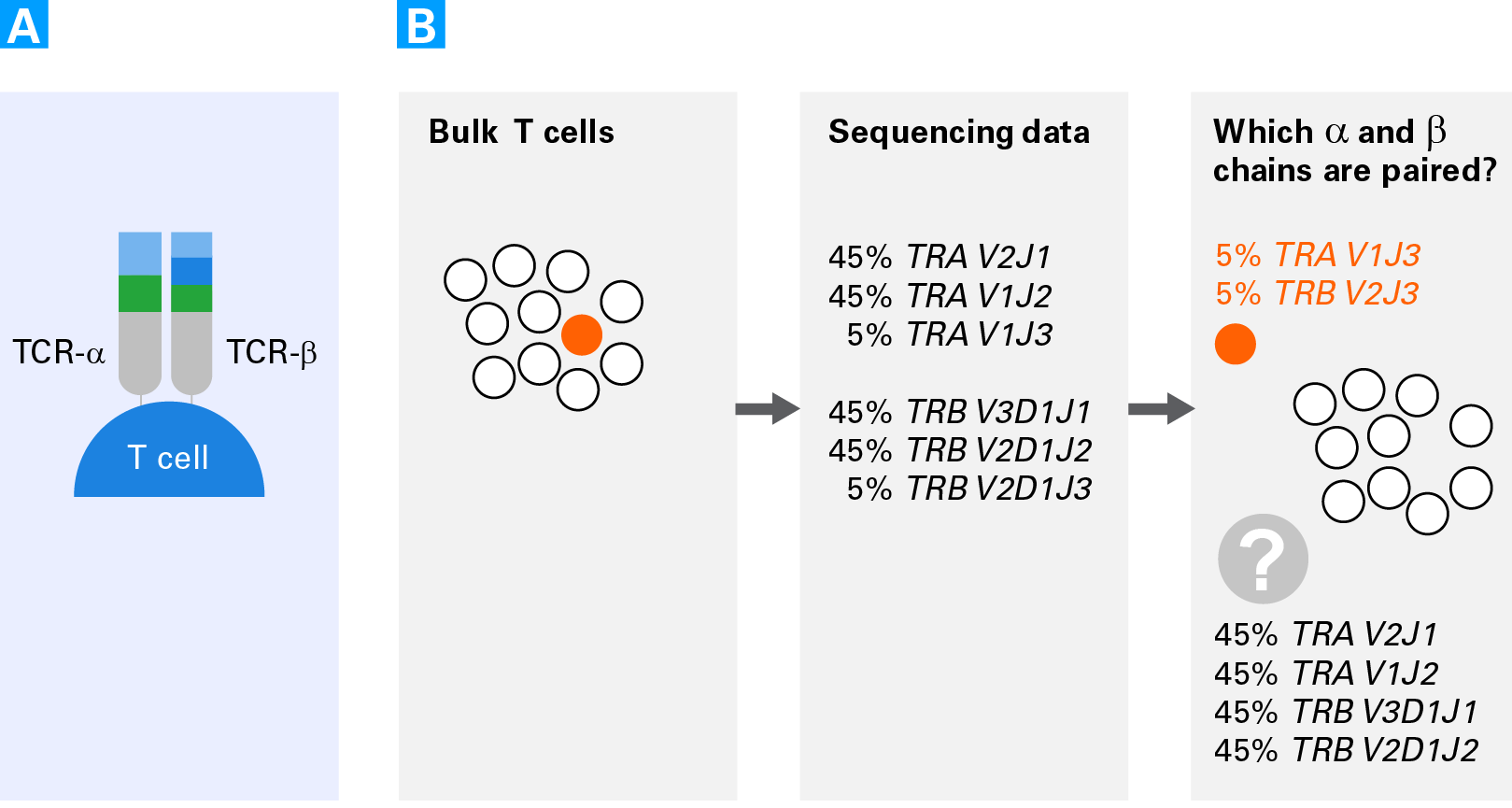
Benefits of studying T cells at the single-cell level. Panel A. Schematic of a T-cell receptor comprised of an alpha chain (TCR-α) and a beta chain (TCR-β). Panel B. Schematic representing the difficulties of obtaining pairing information for alpha and beta chains from bulk sequencing data. Rare clonotypes can allow for assessing the pairing of some cells (clonotypes in orange). However, for the more highly expressed clonotypes (TCRA V2J1, TCRA V1J2, TCRB V3D1J1, TCRB V2D1J2) there are four possible pairing combinations.
Bioanalyzer traces from example TCR libraries
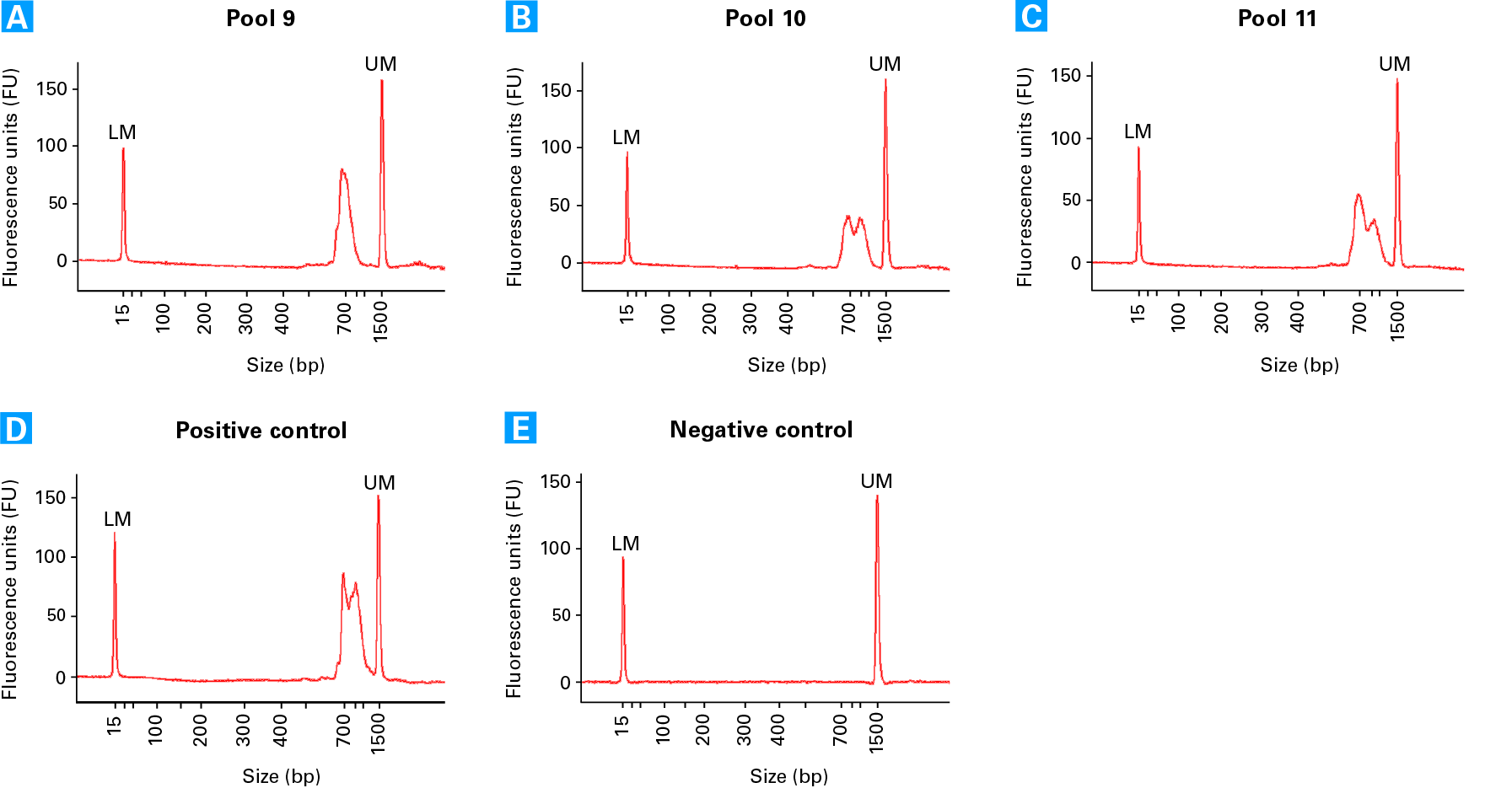
Electropherogram profiles of TCR sequencing libraries. Electropherograms from three different cell pools (see technical note for more details): Pool 9 (Panel A), 10 (Panel B), and 11 (Panel C). The variation in the profiles reflects the abundance of amplified TCRa (peak at ~900 bp) and TCRb (peak at ~700 bp). Panel D. Positive Control RNA TCR library (generated with 8 x 5 pg Control Jurkat Total RNA). Panel E. Negative Control TCR library shows no library is produced.
The SMARTer scTCR workflow uses optimized indexing to allow for pooling 96 cells into 12 libraries.
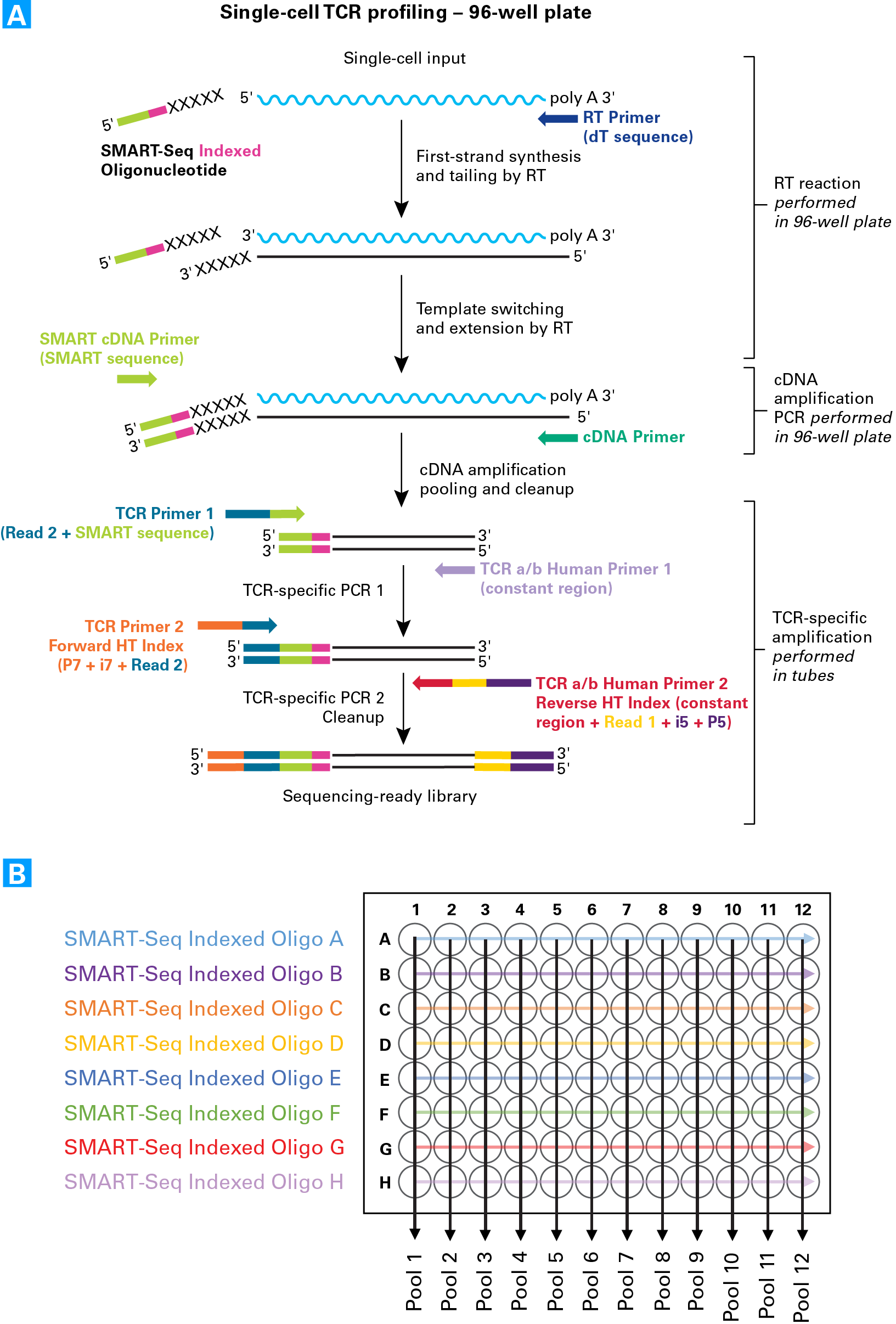
SMARTer Human scTCR a/b Profiling Kit workflow and pooling strategy. Panel A. First-strand cDNA synthesis is dT-primed (RT Primer) and performed by an MMLV-derived reverse transcriptase (RT), which adds nontemplated nucleotides to the 5' end of each mRNA template. The SMART-Seq Indexed Oligos anneal to these nontemplated nucleotides and serve as a template for the incorporation of an additional sequence of nucleotides into the first-strand cDNA by the RT (this is the template-switching step). Each of the eight different SMART-Seq Indexed Oligos provided in the kit each contains a unique six-base in-line index that serves as a cell barcode to allow downstream cell identification after pooling. The additional sequence added to the cDNA by the RT—referred to as the "SMART sequence"—serves as a primer-annealing site for subsequent rounds of PCR, ensuring that only sequences from full-length cDNAs undergo amplification. After pooling (described in Panel B) and a cleanup step, two rounds of gene-specific PCR are performed in succession to amplify cDNA sequences corresponding to variable regions of TCRa and/or TCRb transcripts. The first gene-specific PCR uses the amplified double-stranded cDNA as a template and includes a forward primer with complementarity to the SMART sequence—which also incorporates the Illumina Read 2 sequence (TCR Primer 1)—and reverse primers that are complementary to the constant (i.e., nonvariable) region of TCRa and TCRb (TCR a/b Human Primer 1). The second round of PCR takes the product from the first PCR as a template and uses a forward primer that binds to the Read 2 sequence added by the previous PCR step. The reverse primers bind in the constant region, internal to the PCR1 primers (TCR a/b Human Primer 2 Reverse HT Index) allowing amplification of the entire variable region and a portion of the constant region of TCRa and TCRb cDNA. The forward and reverse primers include adapter and index sequences that are compatible with the Illumina sequencing platform and allow for multiplexing of up to 96 samples in a single flow-cell lane. Panel B. Samples are pooled by column, such that each pool contains eight cells each with a differently indexed SMART-Seq Indexed Oligo. Different combinations of the Forward and Reverse HT indexes are used during PCR 2 to allow multiplexing of the samples into a single flow-cell lane (see the User Manual for more details).
Cell-type calling and alpha-beta pairing analysis of a mixed population of cells
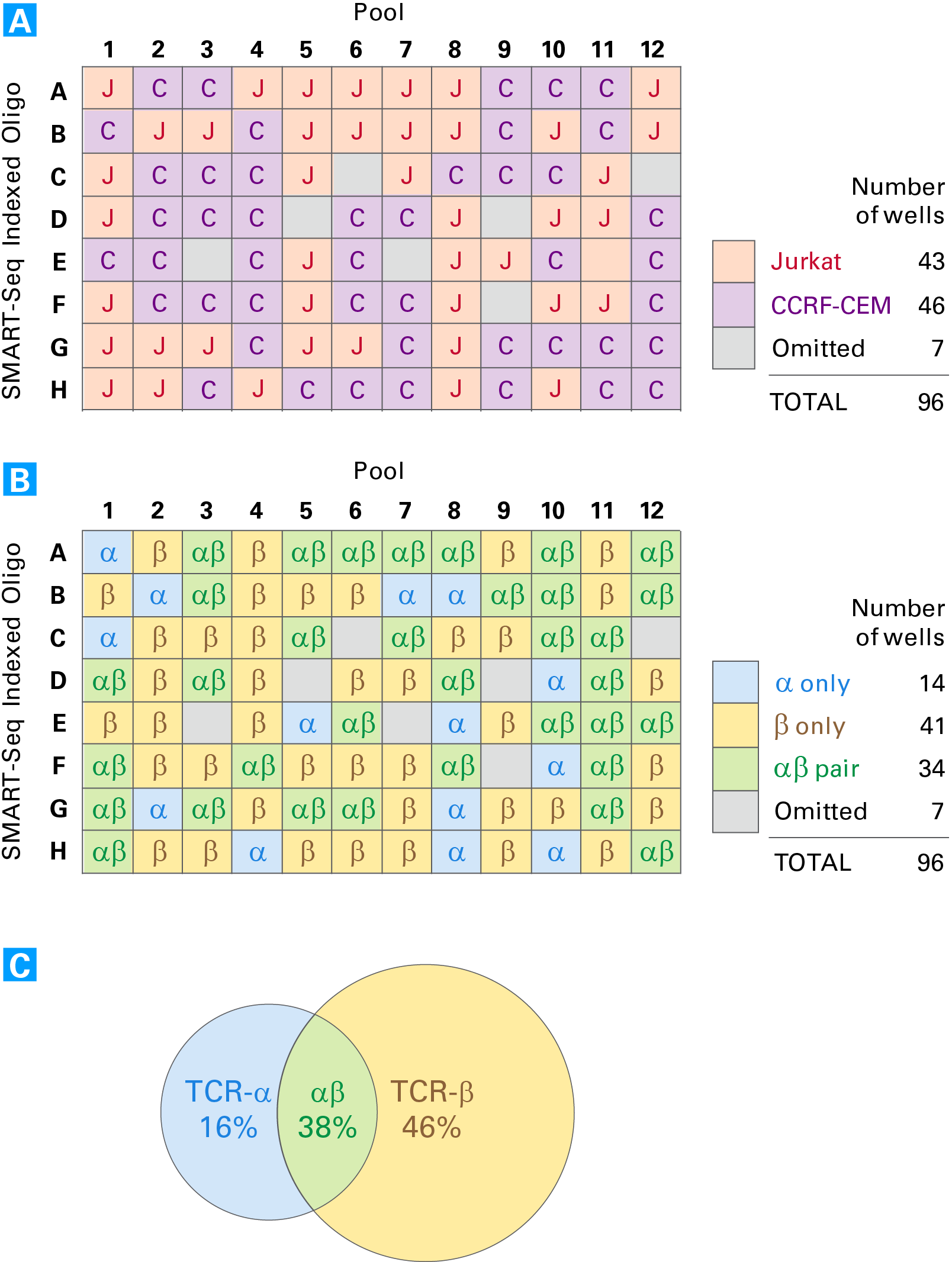
Analysis of a mixed cell population from a 96-well plate. Panel A. Cell-type calling based on the identified clonotypes for each well. The seven omitted cells did not have clonotype calls for either TCRa or TCRb with read numbers that were above the threshold. Panel B. Analysis of pairing information. Paired TCR-αβ chains were obtained for 34 cells in the plate. Panel C. The alpha, beta, or alpha-beta pairing information represented as a percent of cells analyzed. Omitted cells were not included in this analysis.


