Genome-wide sgRNA Library System
Genome-wide sgRNA Library System
Overview
- Contains >76,000 sgRNAs based on the Brunello library which maximizes on-target specificity and activity, while minimizing off-target effects (download sgRNA representation data)
- Each gene in the human genome is targeted by four highly active sgRNAs, enabling screens with a single module
- Modified sgRNA scaffold design for improved Cas9-sgRNA interaction and high editing efficiency
- Lentiviral sgRNA library is already formulated with the Lenti-X Packaging Single Shots and the Xfect Transfection Reagent, such that only water is required to produce a highly efficient transfection mix for transfecting Lenti-X 293T cells to produce high-titer sgRNA libraries
The Guide-it CRISPR Genome-Wide Library PCR Kit is designed for NGS library preparation from cell populations screened using the Guide-it CRISPR Genome-Wide sgRNA Library System (Cat. # 632646), and is also sold as part of the Guide-it CRISPR Genome-Wide sgRNA Library NGS Analysis Kit (Cat. # 632647). The kit contains all the primers and reagents necessary for amplification of proviral sgRNA sequences from screened cell populations for analysis by NGS. This kit includes enough reagents for performing 20 PCRs.
The Guide-it CRISPR Genome-Wide sgRNA Library NGS Analysis Kit enables preparation of next-generation sequencing (NGS) libraries from cell populations screened using the Guide-it CRISPR Genome-Wide sgRNA Library System (Cat. # 632646), allowing for identification of sgRNA sequences and corresponding gene knockouts that have increased or decreased in frequency under the conditions of the screen. The kit provides all the components necessary to prepare ten sequencing libraries (20 PCRs), including a genomic DNA purification kit, primers, PCR enzyme and buffers, and a PCR cleanup kit.
The Guide-it CRISPR Genome-Wide sgRNA Library System includes all of the components necessary to perform five lentiviral-based, genome-wide CRISPR/Cas9 knockout screens in human cells. Provided with the kit are lentiviral vectors for expression of Cas9 and >76,000 sgRNAs targeting >19,000 human genes (four guides per gene) in lyophilized, ready-to-transfect formats that simplify the production of high-titer lentivirus. The sgRNA library included with this kit is based on the Brunello library (Doench et al. 2016), and incorporates design features intended to maximize on-target efficacy and minimize off-target effects. To ensure balanced guide representation, we analyze the sgRNA library by NGS to confirm that >90% of the included sgRNAs fall within a 10-fold distribution range. Enrichment or depletion of sgRNAs in screened cells can be analyzed by NGS with the Guide-it CRISPR Genome-Wide sgRNA Library NGS Analysis Kit (Cat. # 632647).
Overview
- Contains >76,000 sgRNAs based on the Brunello library which maximizes on-target specificity and activity, while minimizing off-target effects (download sgRNA representation data)
- Each gene in the human genome is targeted by four highly active sgRNAs, enabling screens with a single module
- Modified sgRNA scaffold design for improved Cas9-sgRNA interaction and high editing efficiency
- Lentiviral sgRNA library is already formulated with the Lenti-X Packaging Single Shots and the Xfect Transfection Reagent, such that only water is required to produce a highly efficient transfection mix for transfecting Lenti-X 293T cells to produce high-titer sgRNA libraries
Applications
- Pooled genome-wide phenotypic screens
- Loss-of-function genetic screens
Representation of sgRNAs within the Guide-it CRISPR Genome-Wide sgRNA Library System
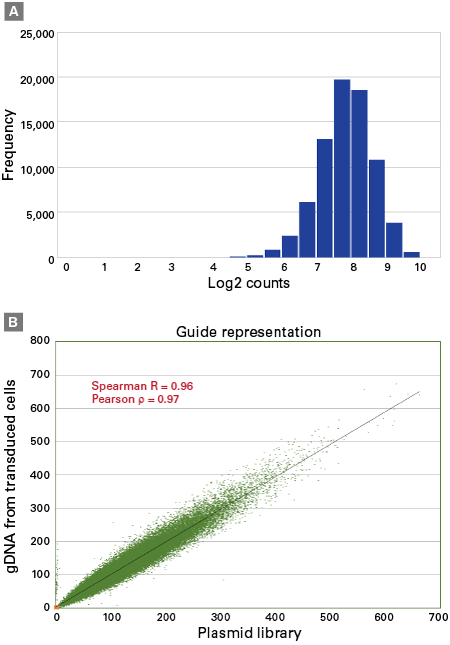
Representation of sgRNAs within the Guide-it CRISPR Genome-Wide sgRNA Library System. Panel A. Guide representation in plasmid library. The Brunello-based sgRNA library was cloned into the pLVXS-sgRNA-mCherry-hyg Vector and amplified. The representation of the sgRNAs within the plasmid DNA library was verified by NGS. The representation of >90% of all the stated sgRNAs within the library was within a 10-fold distribution range. Bars represent the number of sgRNAs with a specific read count. Panel B. Comparison of guide representation in the plasmid library and transduced cells. Genomic DNA was isolated from Cas9+/sgRNA+ A375 cells selected on hygromycin and PCR amplified. The PCR product was sequenced to determine read counts of each integrated sgRNA relative to the starting plasmid DNA population. We observed strong Spearman and Pearson correlations indicating that the system is able to maintain sgRNA representation in the transduced and selected cell population.
Identification and analysis of sgRNAs isolated from cells after a 6-thioguanine screen
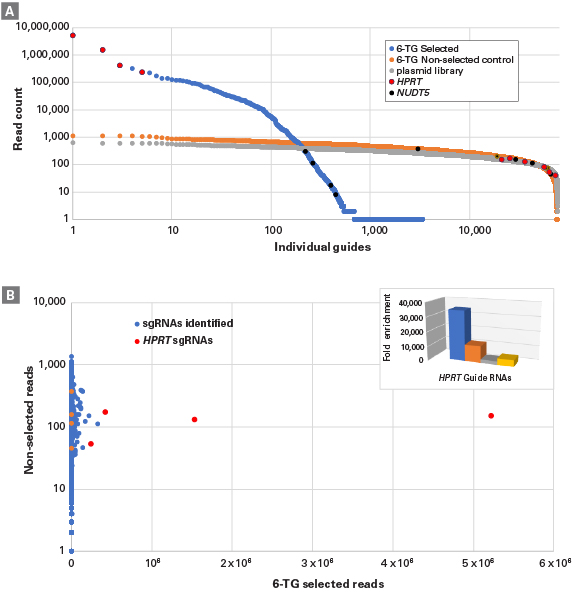
Identification and analysis of sgRNAs isolated from cells after a 6-thioguanine (6-TG) screen. Panel A. sgRNA representation was compared between the original plasmid library population used to make the lentivirus preparation and the resulting transduced, selected and non-selected cell populations. All four sgRNAs for HPRT (red dots) and NUDT5 (black dots) were enriched in the 6-TG-selected population (blue dots). The representation of these sgRNAs in the plasmid library (gray) and transduced, non-selected population (orange) is also shown. Panel B. After selection, the sgRNA representation was shifted in response (blue) to 6-TG. Individual sgRNAs that were enriched are highlighted within this correlation (red and orange dots). Inset shows fold-enrichment of the four sgRNAs for the HPRT gene as compared to the non-selected population.
Determination of transduction efficiency of the sgRNA library lentivirus using mCherry fluorescence
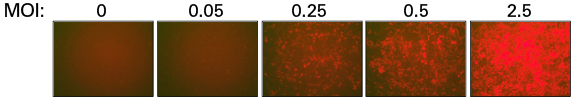
Determination of transduction efficiency of the sgRNA library lentivirus using mCherry fluorescence. Lentivirus containing the sgRNA library was produced following the instructions in the user manual. Lentivirus was harvested after 48 hrs and used to transduce Cas9-expressing A375 cells at varying MOIs. Transduced cells were plated and analyzed for transduction efficiency by fluorescence microscopy and FACS after 48 hours.
Vector and sgRNA scaffold design used in the Guide-it CRISPR sgRNA library
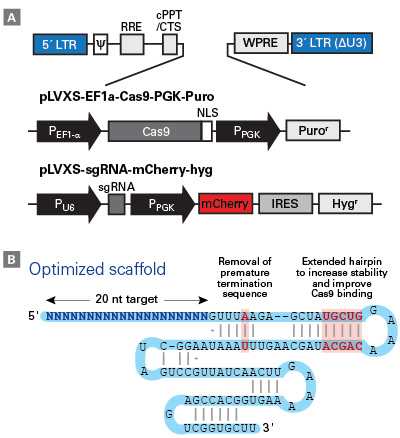
Vector and sgRNA scaffold design used in the Guide-it CRISPR sgRNA library. Panel A. The pLVXS-EF1a-Cas9-PGK-Puro and pLVXS-sgRNA-mCherry-hyg vectors maps showing the lentiviral vector backbone. The vectors are self-inactivating for increased safety during production and use. The pLVXS-sgRNA-mCherry-hyg vector contains both mCherry and hygromycin markers expressed from an IRES-linked bicistronic expression cassette. The sgRNAs are expressed from a human U6 promoter and contain an optimized scaffold sequence for better Cas9 loading and editing efficiency (Panel B).
Workflow schematic for an sgRNA library screen using 6-thioguanine (6-TG) selection
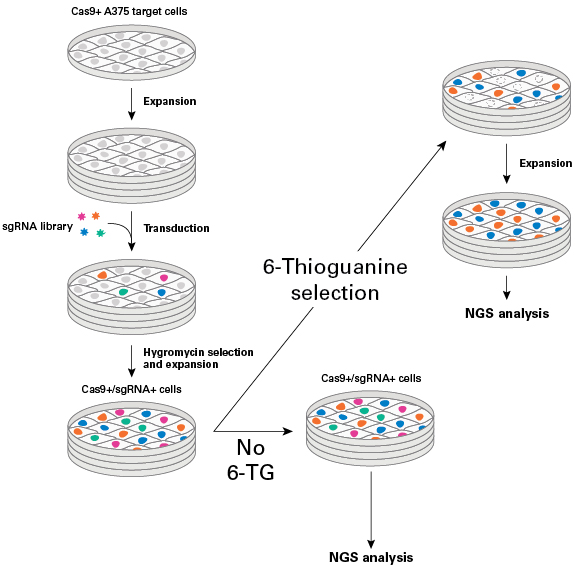
Workflow schematic for an sgRNA library screen using 6-thioguanine (6-TG) selection. After transduction of Cas9+ A375 cells with the sgRNA library, hygromycin selection, and expansion, the Cas9+/ sgRNA+ cells were split into two populations. A part of the Cas9+/sgRNA+ cell population was exposed to 6-TG, while the control population was expanded under normal culture conditions, without adding 6-TG. After expansion of both cell populations, genomic DNA was harvested and prepared for sequencing with the Guide-it CRISPR Genome-Wide sgRNA Library NGS Analysis Kit.
Doench, J.G. Am I ready for CRISPR? A user's guide to genetic screens. Nat. Rev. Genet. 19, 67–80 (2018).
Doench, J.G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).